AI tool for the future precision medicine and precision health.
Based on two PDB structures to simulate any possible contacting sites by torsion angle and coordinate distance, Graph-equivalent models use equivalent attention to learn atom features, position encoding, distance correlations and central tourism angles. Randomly collision energy prediction simulates more possible complexes by estimating contact energy. Models are trained on Protein-Protein and Antibody-Antigen complex data.
In: PDB structure file / Out: Complex PDB file
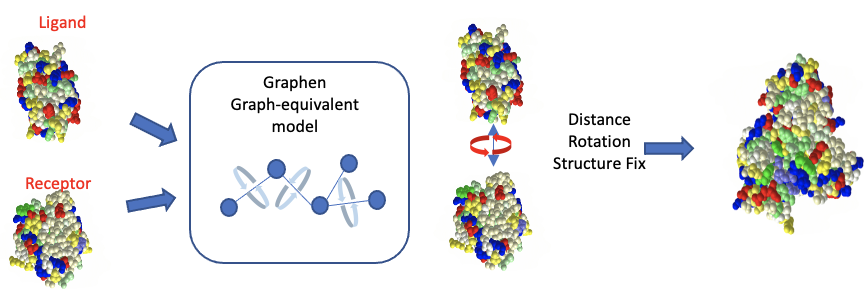
Based on two PDB structures to simulate any possible contacting sites by torsion angle and coordinate distance, Graph-equivalent models use equivalent attention to learn atom features, position encoding, distance correlations and central tourism angles. Randomly collision energy prediction simulates more possible complexes by estimating contact energy. Models are trained on Protein-Protein and Antibody-Antigen complex data.
In: PDB structure file / Out: Complex PDB file
