AI tool for the future precision medicine and precision health.
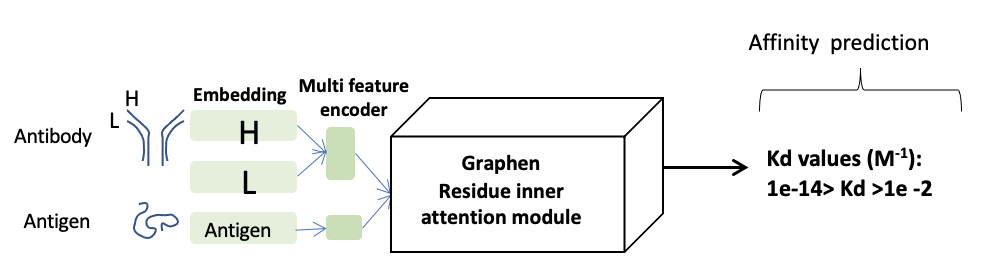
Affinity prediction is a hard-to-determine big molecular interaction. Our prediction model uses pair-wised atom forces and dynamic docking structure to predict the efficacy of affinity, like hydrogen bond, pi-stacking, VDW, etc. Based on Antibody-Antigen interface energy changing, our prediction model is trained by molecular interaction from PDB co-structure complex database, and estimates transform energy to predict Kd/Ka values.
In: Complex PDB file / Out: Affinity score
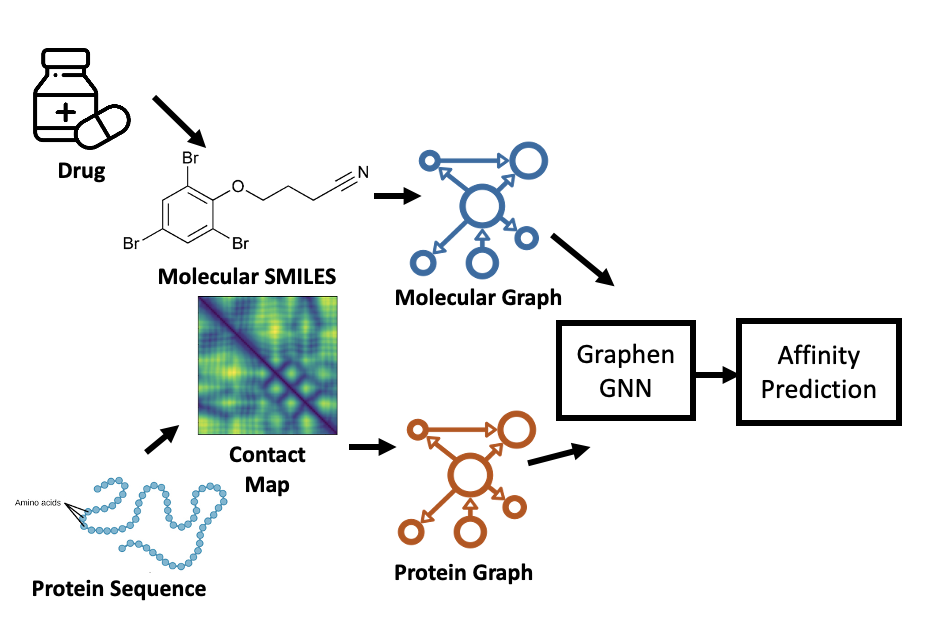
Graphen Drug-and-Target Affinity Prediction tool utilizes Graph Convolutional Network (GCN)-Based molecular structure learning and uses graph representation molecular SMILES encoding and protein structure contact map to find the correlation between drug and target proteins. We use non-Euclidean structure with more efficacy for drug and protein interaction learning. DTA models had been trained from TDC molecular dataset. It predicts the affinity between pharmacophore and protein.
In: PDB file or Fasta / Out: PDB file